Oncology-focused relational dataset derived from MIMIC-IV
This project presents a curated, oncology-focused relational dataset derived from the MIMIC-IV v3.1 clinical database, one of the largest publicly available collections of longitudinal, patient-level hospital data from the Beth Israel Deaconess Medical Center.
While MIMIC-IV is not an oncology-specific cohort and contains a limited number of cancer patients relative to dedicated registries, it provides a rich clinical environment for exploring how oncology-relevant data can be structured, queried, and integrated at scale.
The dataset isolates and organises clinical signals relevant to cancer patients to support exploratory analysis, visualisation, and methodological machine-learning workflows. It is designed as a technical foundation for bridging routine clinical data (MIMIC-IV) with external molecular resources (e.g. TCGA).
- Extracted key tables from MIMIC-IV (
patients,admissions,diagnoses_icd,icustays,prescriptions,labevents) into a local PostgreSQL environment - Filtered all data to focus on oncology-relevant signals:
- Diagnoses using cancer-related ICD-9 (140–239) and ICD-10 (C00–D49) codes
- Laboratory values including blood counts, liver enzymes, and selected tumour markers
- Medications relevant to chemotherapy, immunotherapy, or hormonal therapy
- Defined a derived oncology cohort using ICD-based logic for efficient filtering
- Built supporting views for treatment timelines and labelled laboratory values
- Optimised for interactive querying via indexes and materialised views
Metadata
Metadata
- Source: MIMIC-IV v3.1
- Institution: Beth Israel Deaconess Medical Center (2008–2022)
- Modules Used:
hosp,icu - Programming: PostgreSQL, Python (ETL), SQL (views and indexing)
- Focus: Oncology-relevant EHR subset
- Primary Cohort Definition: Patients with ICD-9/10 neoplasm codes
- Compliance: PhysioNet DUA 1.5.0, HIPAA-deidentified, time-shifted
Core Schema Structure
Schema Structure
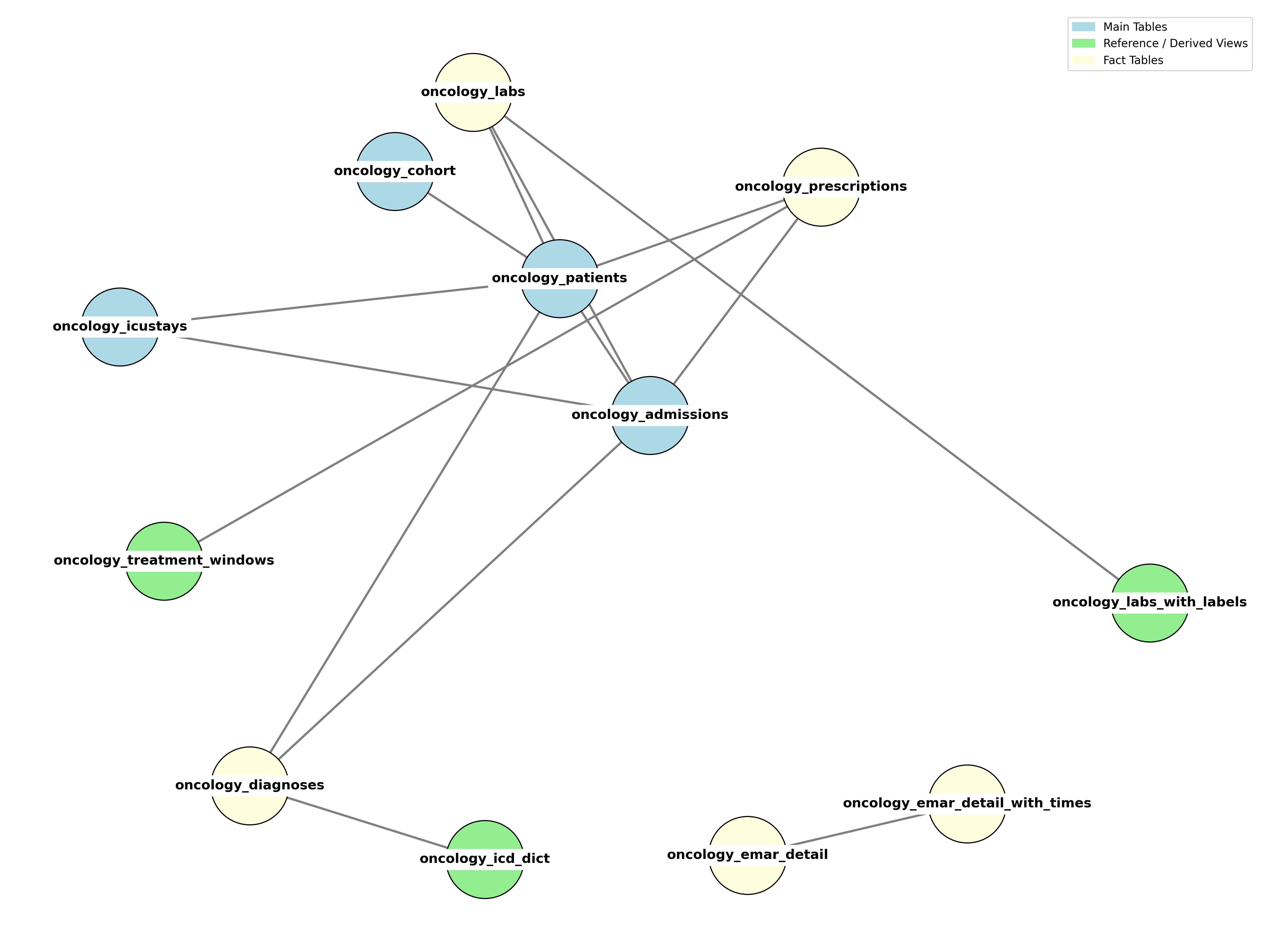
The dataset is organised into three complementary table categories:
| Category | Description | Examples |
|---|---|---|
| Main Tables | Core entities: patients, admissions, ICU stays, cohort flags | oncology_patients, oncology_admissions, oncology_icustays, oncology_cohort |
| Fact Tables | Event-level clinical records: diagnoses, labs, prescriptions, medication admin | oncology_diagnoses, oncology_labs, oncology_prescriptions, oncology_emar_detail |
| Reference Tables | Dictionaries and derived views for enrichment and summarisation | oncology_icd_dict, oncology_labs_with_labels, oncology_emar_detail_with_times, oncology_treatment_windows |
Use Cases
- Track cancer patients longitudinally (laboratory values, medications, ICU exposure)
- Prototype outcome or response modelling pipelines (methodological scope)
- Build dashboards and exploratory clinical analytics
- Practice reproducible clinical data engineering and SQL workflows
- Integrate clinical trajectories with external genomics resources (e.g. TCGA)