Integrating Kinase Bioactivity with Expression Profiles
Here I showcase a translational knowledge graph that integrates reference datasets using semantic web technologies to support cross-domain questions in oncology target discovery.
As a proof of concept, it connects:
- Kinase-targeted bioactivity (ChEMBL; compound–target potency and assay context)
- Normal tissue expression (GTEx)
- Tumour expression (TCGA)
- Identifier harmonisation (UniProt-centered mappings to Ensembl and anatomy terms)
All data are represented as RDF and loaded into a GraphDB triple store as named graphs, enabling reproducible semantic joins across datasets without pre-materialising every combination.
Pipeline Overview
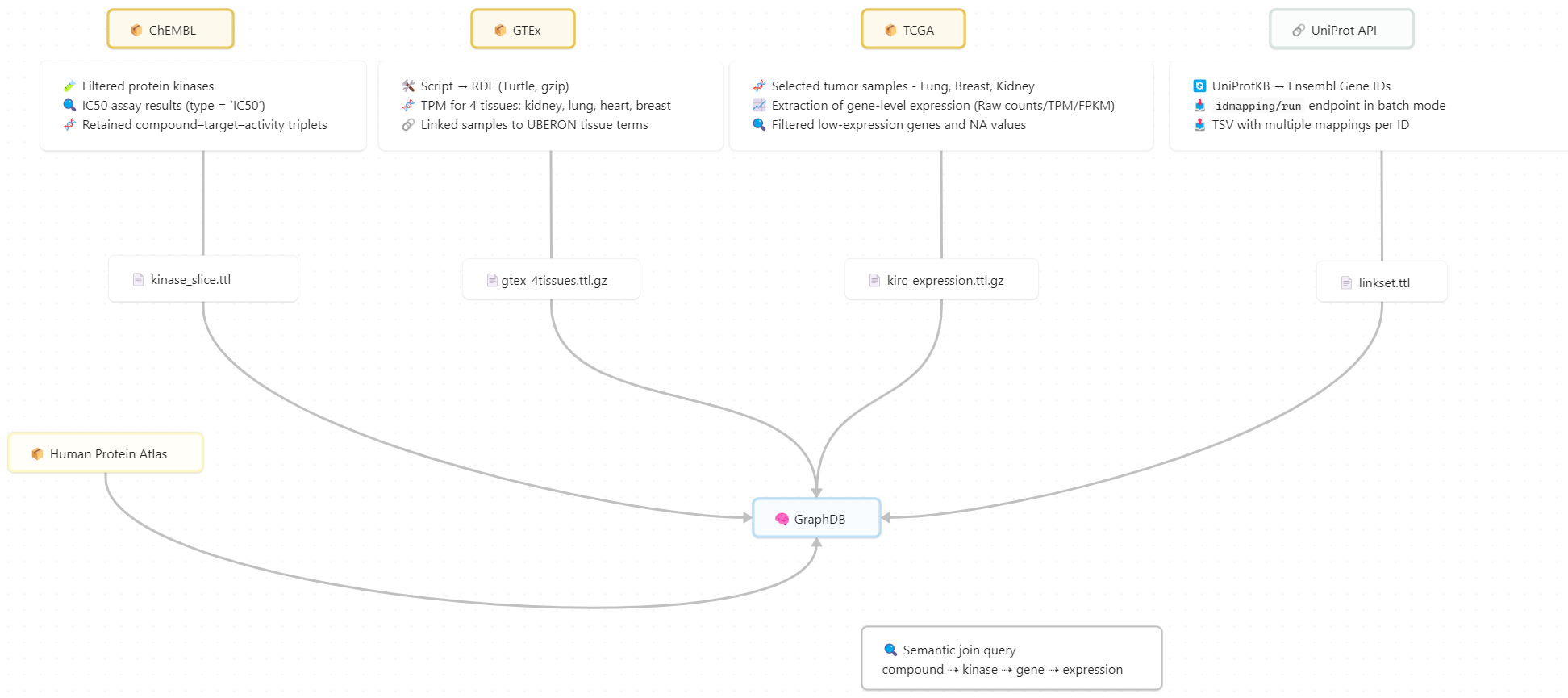
Each source dataset (ChEMBL, GTEx, TCGA, UniProt) is processed independently and exported to Turtle (TTL), then loaded into GraphDB as separate named graphs. Datasets are harmonised through ontology-aligned identifiers (e.g. UniProt ↔ Ensembl; anatomy terms such as UBERON) to make cross-dataset joins explicit and inspectable.
Compound → Kinase Target → Gene → Expression (GTEx / TCGA)
This enables questions such as:
- "Which compounds inhibit kinases whose genes are overexpressed in kidney tumours but not in healthy kidney cortex?"
- "Which kinase targets show tumour-specific expression patterns across multiple tissues?"
The Human Protein Atlas is included as a planned extension to add protein-level validation and reduce reliance on transcript-only signals.
Use Cases
This integrated knowledge graph supports semantic exploration across compounds, kinase targets, gene identifiers, and expression profiles in normal and tumour tissues. Example use cases include:
- Prioritise kinase targets using tumour-specific overexpression signals
- De-risk targets by filtering for low expression in critical healthy tissues (toxicity proxy)
- Rank compounds by the expression of their target set in a selected cancer type
- Compare expression contrast across tissues (e.g. GTEx vs. TCGA) to identify specificity
- Assess compound–target coverage across tumour types for combination or repositioning strategies